AI: algorithms improve the analysis of medical images
Artificial intelligence can improve the analysis of medical image data. For example, algorithms based on deep learning can determine the location and size of tumors. This is the result of autoPET, an international competition for medical image analysis. Researchers from the Karlsruhe Institute of Technology (KIT) took fifth place. The seven best autoPET teams report in the journal Nature Machine Intelligence (DOI: 10.1038/s42256-024-00912-9) on how algorithms can detect tumor lesions in positron emission tomography (PET) and computed tomography (CT).
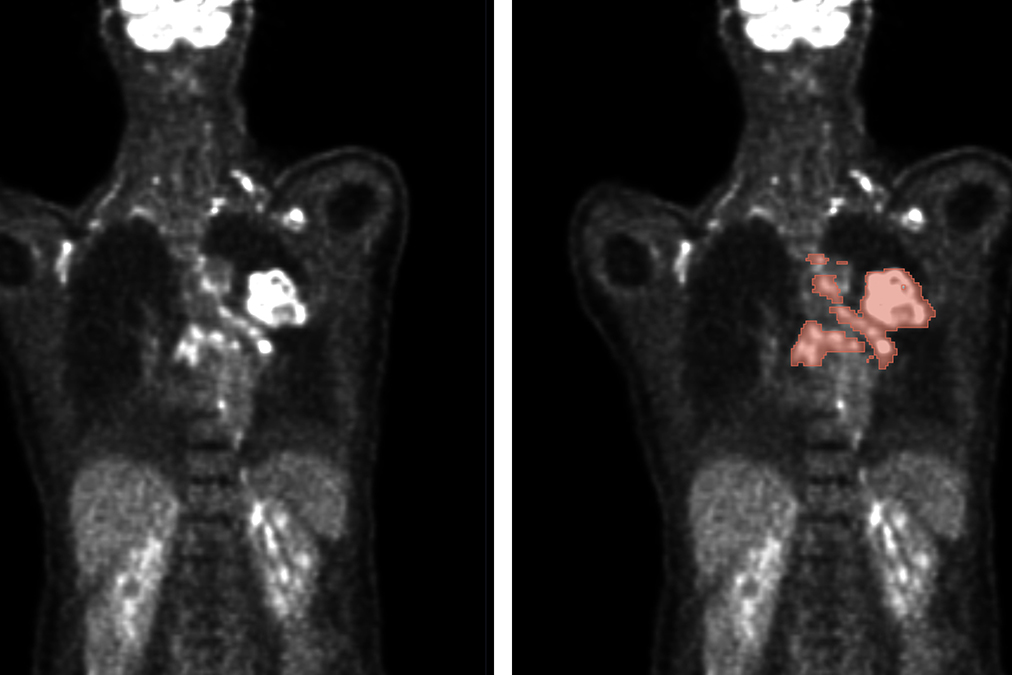
Automated methods enable the analysis of PET/CT scans (left) to accurately predict tumor location and size (right) for improved diagnosis and treatment planning. (Figure: Gatidis S, Kuestner T. (2022) A whole-body FDG-PET/CT dataset with manually annotated tumor lesions (FDG-PET-CT-Lesions) [Dataset]. The Cancer Imaging Archive. DOI: 10.7937/gkr0-xv29)
Imaging techniques play a key role in the diagnosis of cancer. Precisely determining the location, size and type of tumor is crucial to finding the right therapy. The most important imaging techniques include positron emission tomography (PET) and computer tomography (CT). PET uses radionuclides to visualize metabolic processes in the body. Malignant tumors often have a much more intensive metabolism than benign tissue. Radioactively labeled glucose, usually fluorine-18-deoxyglucose (FDG), is used for this purpose. In CT, the body is scanned layer by layer in an X-ray tube to visualize the anatomy and localize tumours.
Automation can save time and improve evaluation
Cancer patients sometimes have hundreds of lesions, i.e. pathological changes caused by the growth of tumors. To obtain a uniform picture, all lesions must be recorded. Doctors determine the size of the tumor lesions by manually marking 2D slice images - an extremely time-consuming task. "Automated evaluation using an algorithm would save an enormous amount of time and improve the results," explains Professor Rainer Stiefelhagen, Head of the Computer Vision for Human-Computer Interaction Lab (cv:hci) at KIT.
Rainer Stiefelhagen and Zdravko Marinov, a doctoral student at cv:hci, took part in the international autoPET competition in 2022 and came fifth out of 27 teams with 359 participants from all over the world. The Karlsruhe researchers formed a team with Professor Jens Kleesiek and Lars Heiliger from the IKIM - Institute for Artificial Intelligence in Medicine in Essen. Organized by the University Hospital Tübingen and the LMU Klinikum München, autoPET combined imaging and machine learning. The task was to automatically segment metabolically active tumor lesions on whole-body PET/CT. For the algorithm training, the participating teams had access to a large annotated PET/CT dataset. All algorithms submitted for the final phase of the competition are based on deep learning methods. This is an area of machine learning that uses multi-layered artificial neural networks to recognize complex patterns and correlations in large amounts of data. The seven best teams from the autoPET competition have now reported on the possibilities of automated analysis of medical image data in the journal Nature Machine Intelligence.
Algorithm ensemble best detects tumor lesions
As the researchers explain in their publication, an ensemble of the top-rated algorithms proved to be superior to individual algorithms. The ensemble of algorithms can detect tumor lesions efficiently and precisely. "However, the performance of the algorithms in image data evaluation depends on the quantity and quality of the data, but also on the algorithm design, for example with regard to the decisions made in the post-processing of the predicted segmentation," explains Professor Rainer Stiefelhagen. Further research is needed to improve the algorithms and make them more robust against external influences so that they can be used in everyday clinical practice. The aim is to fully automate the analysis of medical image data from PET and CT in the near future.
Original publication
Sergios Gatidis, Marcel Früh, Matthias P. Fabritius, Sijing Gu, Konstantin Nikolaou, Christian La Fougère, Jin Ye, Junjun He, Yige Peng, Lei Bi, Jun Ma, Bo Wang, Jia Zhang, Yukun Huang, Lars Heiliger, Zdravko Marinov, Rainer Stiefelhagen, Jan Egger, Jens Kleesiek, Ludovic Sibille, Lei Xiang, Simone Bendazzoli, Mehdi Astaraki, Michael Ingrisch, Clemens C. Cyran & Thomas Küstner: Results from the autoPET challenge on fully automated lesion segmentation in oncologic PET/CT imaging. Nature Machine Intelligence, 2024. DOI: 10.1038/s42256-024-00912-9
